Abstract
Results of the ISMRM 2020 joint Reproducible Research & Quantitative MR study groups reproducibility challenge on phantom and human brain T1 mapping
*Mathieu Boudreau1,2, *Agah Karakuzu1, Julien Cohen-Adad1,3,4,5, Ecem Bozkurt6, Madeline Carr7,8, Marco Castellaro9, Luis Concha10, Mariya Doneva11, Seraina Dual12, Alex Ensworth13,14, Alexandru Foias1, Véronique Fortier15,16, Refaat E. Gabr17, Guillaume Gilbert18, Carri K. Glide-Hurst19, Matthew Grech-Sollars20,21, Siyuan Hu22, Oscar Jalnefjord23,24, Jorge Jovicich25, Kübra Keskin6, Peter Koken11, Anastasia Kolokotronis13,26, Simran Kukran27,28, Nam. G. Lee6, Ives R. Levesque13,29, Bochao Li6, Dan Ma22, Burkhard Mädler30, Nyasha Maforo31,32, Jamie Near33,34, Erick Pasaye35, Alonso Ramirez-Manzanares36, Ben Statton37,Christian Stehning30, Stefano Tambalo25, Ye Tian6, Chenyang Wang38, Kilian Weiss30, Niloufar Zakariaei39, Shuo Zhang30, Ziwei Zhao6, Nikola Stikov1,2,40
- *Authors MB and AK contributed equally to this work
1NeuroPoly Lab, Polytechnique Montréal, Montreal, Quebec, Canada, 2Montreal Heart Institute, Montreal, Quebec, Canada, 3Unité de Neuroimagerie Fonctionnelle (UNF), Centre de recherche de l’Institut Universitaire de Gériatrie de Montréal (CRIUGM), Montreal, Quebec, Canada, 4Mila - Quebec AI Institute, Montreal, QC, Canada, 5Centre de recherche du CHU Sainte-Justine, Université de Montréal, Montreal, QC, Canada, 6Magnetic Resonance Engineering Laboratory (MREL), University of Southern California, Los Angeles, California, USA, 7Medical Physics, Ingham Institute for Applied Medical Research, Liverpool, Australia, 8Department of Medical Physics, Liverpool and Macarthur Cancer Therapy Centres, Liverpool, Australia, 9Department of Information Engineering, University of Padova, Padova, Italy, 10Institute of Neurobiology, Universidad Nacional Autónoma de México Campus Juriquilla, Querétaro, México, 11Philips Research Hamburg, Germany, 12Stanford University, Stanford, California, United States, 13Medical Physics Unit, McGill University, Montreal, Canada, 14University of British Columbia, Vancouver, Canada, 15Department of Medical Imaging, McGill University Health Centre, Montreal, Quebec, Canada 16Department of Radiology, McGill University, Montreal, Quebec, Canada, 17Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center at Houston, McGovern Medical School, Houston, Texas, USA, 18MR Clinical Science, Philips Canada, Mississauga, Ontario, Canada, 19Department of Human Oncology, University of Wisconsin-Madison, Madison, Wisconsin, USA, 20Centre for Medical Image Computing, Department of Computer Science, University College London, London, UK, 21Lysholm Department of Neuroradiology, National Hospital for Neurology and Neurosurgery, University College London Hospitals NHS Foundation Trust, London, UK, 22Department of Biomedical Engineering, Case Western Reserve University, Cleveland, Ohio, USA, 23Department of Medical Radiation Sciences, Institute of Clinical Sciences, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 24Biomedical Engineering, Sahlgrenska University Hospital, Gothenburg, Sweden, 25Center for Mind/Brain Sciences, University of Trento, Italy, 26Hopital Maisonneuve-Rosemont, Montreal, Canada, 27Bioengineering, Imperial College London, UK, 28Radiotherapy and Imaging, Insitute of Cancer Research, Imperial College London, UK, 29Research Institute of the McGill University Health Centre, Montreal, Canada, 30Clinical Science, Philips Healthcare, Germany, 31Department of Radiological Sciences, University of California Los Angeles, Los Angeles, CA, USA, 32Physics and Biology in Medicine IDP, University of California Los Angeles, Los Angeles, CA, USA, 33Douglas Brain Imaging Centre, Montreal, Canada, 34Sunnybrook Research Institute, Toronto, Canada, 35National Laboratory for Magnetic Resonance Imaging, Institute of Neurobiology, Universidad Nacional Autónoma de México (UNAM), Juriquilla, Mexico, 36Computer Science Department, Centro de Investigación en Matemáticas, A.C., Guanajuato, México, 37Medical Research Council, London Institute of Medical Sciences, Imperial College London, London, United Kingdom, 38Department of Radiation Oncology - CNS Service, The University of Texas MD Anderson Cancer Center, Texas, USA, 39Department of Biomedical Engineering, University of British Columbia, British Columbia, Canada, 40Center for Advanced Interdisciplinary Research, Ss. Cyril and Methodius University, Skopje, North Macedonia
Abstract#
Purpose: T1 mapping is a widely used quantitative MRI technique, but its tissue-specific values remain inconsistent across protocols, sites, and vendors. The ISMRM Reproducible Research study group (RRSG) and Quantitative MR study group (qMRSG) jointly launched a T1 mapping reproducibility challenge. The goal was to investigate whether an independently-implemented image acquisition protocol at multiple centers could reliably measure T1 using inversion recovery in a standardized phantom and in brains of healthy volunteers.
Methods: The reproducibility of a robust T1 mapping protocol and fitting algorithm from a well-known Magnetic Resonance in Medicine publication (Barral et al. 2010) were evaluated. Participants in the challenge were invited to acquire T1 mapping data on a standard ISMRM/NIST phantom and/or in healthy human brains. Data submission, pipeline development, and analysis were performed using open-source platforms for increased reproducibility and transparency. For participants acquiring data at multiple sites, inter-submission and intra-submission comparisons were conducted by selecting one dataset per submission.
Results: Eighteen submissions were accepted using data collected from MRIs from three manufacturers (Siemens, GE, Philips) and at 3T (except one submission, at 0.35T), resulting in a total of 38 datasets acquired in phantoms and 56 datasets in healthy human subjects. Inter-submission mean coefficient of variation (CoV) was 6.1% for phantom measurements, nearly twice as high as the evaluated intra-submission CoV (2.9%). A similar trend was observed in the human data (inter-/intra- submission CoV was 6.0/2.9 % in the genu and 16/6.9 % in the cortical gray matter).
Conclusion: The imaging protocol and fitting algorithm from Barral et al. (2010) resulted in good reproducibility of both phantom and human brain T1 measurements. Differences in implementations between submissions resulted in higher variance in reported values relative to intra-submission variability. This challenge generated a large open database of inversion recovery T1 mapping acquisitions across several sites and MRI vendors accessible at osf.io/ywc9g/. To allow for the wider community to access and engage with this dataset, we developed an interactive dashboard that is available at https://rrsg2020.dashboards.neurolibre.org.
Dashboard: Challenge Submissions
1 | INTRODUCTION#
Significant challenges exist in the reproducibility of quantitative MRI (qMRI) [Keenan et al., 2019]. Despite its promise of improving the specificity and reproducibility of MRI acquisitions, few qMRI techniques have been integrated into clinical practice. Even the most fundamental MR parameters cannot be measured with sufficient reproducibility and precision across clinical scanners to pass the second of six stages of technical assessment for clinical biomarkers [Fryback and Thornbury, 1991, Schweitzer, 2016, Seiberlich et al., 2020]. Half a century has passed since the first quantitative T1 (spin-lattice relaxation time) measurements were first reported as a potential biomarker for tumors [Damadian, 1971], followed shortly thereafter by the first in vivo quantitative T1 maps [Pykett and Mansfield, 1978] of tumors, but there is still disagreement in reported values for this fundamental parameter across different sites, vendors, and implementations [Stikov et al., 2015].
Among fundamental MRI parameters, T1 holds significant importance [Boudreau et al., 2020]. It represents the time it takes for the longitudinal magnetization to recover after being disturbed by an RF pulse. The T1 value varies based on molecular mobility and magnetic field strength [Bottomley et al., 1984, Dieringer et al., 2014, Wansapura et al., 1999], making it a valuable parameter for distinguishing different tissue types. Accurate knowledge of T1 values is essential for optimizing clinical MRI pulse sequences for contrast and time efficiency [Ernst and Anderson, 1966, Redpath and Smith, 1994, Tofts, 1997] and as a calibration parameter for other quantitative MRI techniques [Sled and Pike, 2001, Yuan et al., 2012]. Among the number of techniques to measure T1, inversion recovery (IR) [Drain, 1949, Hahn, 1949] is widely held as the gold standard technique, as it is robust against other effects (e.g. B1 inhomogeneity) and potential errors in measurements (e.g. insufficient spoiling) [Stikov et al., 2015]. However, because the technique requires a long repetition time (TR > T1), it is very slow and impractical for whole-organ measurements, limiting its clinical use. In practice, it is mostly used as a reference to validate other T1 mapping techniques, such as variable flip angle imaging (VFA) [Cheng and Wright, 2006, Deoni et al., 2003, Fram et al., 1987], Look-Locker [Look and Locker, 1970, Messroghli et al., 2004, Piechnik et al., 2010], and MP2RAGE [Marques and Gruetter, 2013, Marques et al., 2010].
Efforts have been made to develop quantitative MRI phantoms to assist in standardizing T1 mapping methods [Keenan et al., 2018]. A quantitative MRI standard system phantom was created in a joint project between the International Society for Magnetic Resonance in Medicine (ISMRM) and the National Institute of Standards and Technology (NIST) [Stupic et al., 2021], and has since been commercialized (Premium System Phantom, CaliberMRI, Boulder, Colorado). The spherical phantom has a 57-element fiducial array containing spheres with doped liquids that model a wide range of T1, T2, and PD values. The reference values of each sphere were measured using NMR at 3T [Stupic et al., 2021]. The standardized concentration for relaxometry values established as references by NIST are also used by another company for their relaxometry MRI phantoms (Gold Standard Phantoms Ltd., Rochester, England). The cardiac TIMES phantom [Captur et al., 2016] is another commercially available system phantom focusing on T1 and T2 values in blood and heart muscles, pre- and post-contrast. The ISMRM/NIST phantom has been used in several large multicenter studies already, for example in [Bane et al., 2018] where they compared measurements at eight sites on a single ISMRM/NIST phantom using the inversion recovery and VFA T1 mapping protocols recommended by NIST, as well as some site-specific imaging protocols used for dynamic contrast enhanced (DCE) imaging. Bane et al. [2018] concluded that the acquisition protocol, field strength, and T1 value of the sample impacted the level of accuracy, repeatability, and interplatform reproducibility that was observed. In another study led by NIST researchers [Keenan et al., 2021], T1 measurements were done at two clinical field strengths (1.5T and 3.0 T) and 27 MRI systems (three vendors) using the recommended NIST protocols. That study, which only investigated phantoms, found no significant relationship between T1 discrepancies of the measurements and the MRI vendors used.
The 2020 ISMRM reproducibility challenge 1 posed the following question:
Will an imaging protocol independently-implemented at multiple centers reliably measure what is considered one of the fundamental MR parameters (T1) using the most robust technique (inversion recovery) in a standardized phantom (ISMRM/NIST system phantom) and in the healthy human brain?
More broadly, this challenge aimed at assessing the reproducibility of a qMRI method presented in a seminal paper, [Barral et al., 2010], by evaluating the variability in measurements observed by different research groups that implemented this imaging protocol. As the focus of this challenge was on reproducibility, the challenge design emphasized the use of reproducible research practices, such as sharing code, pipelines, data, and scripts to reproduce figures.
2 | METHODS#
2.1 | Phantom and human data#
The phantom portion of the challenge was launched for those with access to the ISMRM/NIST system phantom [Stupic et al., 2021] (Premium System Phantom, CaliberMRI, Boulder, Colorado). Two versions of the phantom have been produced with slightly different T1/T2/PD values in the liquid spheres, and both versions were used in this study. Phantoms with serial numbers 0042 or less are referred to as “Version 1”, and those with 0043 or greater are “Version 2”. The phantom has three plates containing sets of 14 spheres for ranges of proton density (PD), T1 (NiCl2), and T2 (MnCl2) values. Reference T1 values at 20 °C and 3T for the T1 plate are listed in Table 1 for both versions of the phantom. Researchers that participated in the challenge were instructed to record the temperature before and after scanning the phantom using the phantom’s internal thermometer. Instructions for positioning and setting up the phantom were devised by NIST after they had designed the phantom (prior to the challenge), and were provided to participants through the NIST website 2. In brief, instructions included details about how to orient the phantom consistently at different sites, and how long the phantom should be in the scanner room prior to scanning so that a thermal equilibrium was achieved prior to scanning.
Sphere |
Version 1 (ms) |
Version 2 (ms) |
|---|---|---|
1 |
1989 ± 1.0 |
1883.97 ± 30.32 |
2 |
1454 ± 2.5 |
1330.16 ± 20.41 |
3 |
984.1 ± 0.33 |
987.27 ± 14.22 |
4 |
706 ± 1.0 |
690.08 ± 10.12 |
5 |
496.7 ± 0.41 |
484.97 ± 7.06 |
6 |
351.5 ± 0.91 |
341.58 ± 4.97 |
7 |
247.13 ± 0.086 |
240.86 ± 3.51 |
8 |
175.3 ± 0.11 |
174.95 ± 2.48 |
9 |
125.9 ± 0.33 |
121.08 ± 1.75 |
10 |
89.0 ± 0.17 |
85.75 ± 1.24 |
11 |
62.7 ± 0.13 |
60.21 ± 0.87 |
12 |
44.53 ± 0.090 |
42.89 ± 0.44 |
13 |
30.84 ± 0.016 |
30.40 ± 0.62 |
14 |
21.719 ± 0.005 |
21.44 ± 0.31 |
Participants were also instructed to collect T1 maps in the brains of healthy human participants, if possible. To ensure consistency across datasets, single-slice positioning parallel to the anterior commissure - posterior commissure (AC-PC) line was recommended. Before the scanning process, the participants granted their consent 4 to share their de-identified data openly with the challenge organizers and on the website Open Science Framework (OSF.io). As the submitted single-slice inversion recovery images would be along the AC-PC line, they are unlikely to contain sufficient information for facial identification, and therefore participants were not instructed to de-face their data. The researchers who submitted human data for the challenge provided written confirmation to the organizers that their data was acquired in accordance with their institutional ethics committee (or equivalent regulatory body) and that the subjects had consented to sharing their data as described in the challenge.
2.2 | Protocol#
Participants were instructed to acquire the T1 mapping data using the spin-echo inversion recovery protocol for T1 mapping as reported in [Barral et al., 2010], and detailed in Table 2. This protocol uses four inversion times optimized for human brain T1 values and uses a relatively short TR (2550 ms). It is important to note that this acquisition protocol is not suitable for T1 fitting models that assume TR > 5T1. Instead, more general models of inversion recovery, such as the Barral et al. [2010] fitting model described in Section 2.4.1, can be used to fit this data.
Pulse sequence |
Spin-echo inversion recovery |
|---|---|
Repetition Time (TR) |
2550ms |
Inversion Time (TI) |
50, 400, 1100, 2500 ms |
Echo Time (TE) |
14 ms |
In-plane resolution |
1x1 mm2 |
Slice thickness |
2mm |
Researchers who participated in the challenge were advised to adhere to this protocol as closely as possible, and to report any differences in protocol parameters due to technical limitations of their scanners and/or software. It was recommended that participants submit complex data (magnitude and phase, or real and imaginary), but magnitude-only data was also accepted if complex data could not be conveniently exported.
2.3 | Data Submissions#
Data submissions for the challenge were managed through a dedicated repository on GitHub, accessible at https://github.com/rrsg2020/data_submission. This allowed transparent and open review of the submissions, as well as standardization of the process. All datasets were converted to the NIfTI file format, and images from different TIs needed to be concatenated into the fourth (or “time”) dimension. Magnitude-only datasets required one NIfTI file, while complex datasets required two files (magnitude and phase, or real and imaginary). Additionally, a YAML (*.yaml) configuration file containing submission, dataset, and acquisition details (such as data type, submitter name and email, site details, phantom or volunteer details, and imaging protocol details) was required for each submitted dataset to ensure that the information was standardized and easily found. Each submission was reviewed to confirm that guidelines were followed, and then datasets and configuration files were uploaded to OSF.io (osf.io/ywc9g). A Jupyter Notebook [Kluyver et al., 2016, Beg et al., 2021] pipeline was used to generate T1 maps using qMRLab [Cabana et al., 2015, Karakuzu et al., 2020] and quality-check the datasets prior to accepting the submissions; in particular, we assured that the NiCL2 array was imaged, that the DICOM images were correctly converted to NIfTI, the each images for each acquired TI were not renormalized (in particular, Philips platforms have different image export options that changes how the images are scaled, and a reconversion was necessary to ensure proper scaling for quantitative imaging in some cases – see the submissions GitHub issue #5 for one example 5) for the purposes of quality assurance. Links to the Jupyter Notebook for reproducing the T1 map were shared using the MyBinder platform in each respective submission GitHub issue, ensuring that computational environments (eg, software dependencies and packages) could be reproduced to re-run the pipeline in a web browser.
2.4 | Data Processing#
2.4.1 | Fitting Model and Pipeline#
A reduced-dimension non-linear least squares (RD-NLS) approach was used to fit the complex general inversion recovery signal equation:
where a and b are complex constants. This approach, introduced in , models the general T1 signal equation without the long-TR approximation. The a and b constants inherently factor TR in them, as well as other imaging parameters (eg, excitation and refocusing flip angles, TE, etc). Barral et al. [2010] shared the implementation of the fitting algorithm used in their paper 6. Magnitude-only data were fitted to a modified-version of [1] (Eq. 15 of Barral et al. [2010]) with signal-polarity restoration. To facilitate its use in our pipelines, a wrapper was implemented around this code available in the open-source software qMRLab [Cabana et al., 2015, Karakuzu et al., 2020], which provides a commandline interface (CLI) to call the fitting in MATLAB/Octave scripts.
A Jupyter Notebook data processing pipeline was written using MATLAB/Octave. This pipeline automatically downloads all the datasets from the data-hosting platform osf.io (osf.io/ywc9g), loads each dataset configuration file, fits the T1 data voxel-wise, and exports the resulting T1 map to the NIfTI and PNG formats. This pipeline is available in a GitHub repository (https://github.com/rrsg2020/t1_fitting_pipeline, filename: RRSG_T1_fitting.ipynb). Once all submissions were collected and the pipeline was executed, the T1 maps were uploaded to OSF (osf.io/ywc9g).
2.4.2 | Image Labeling & Registration#
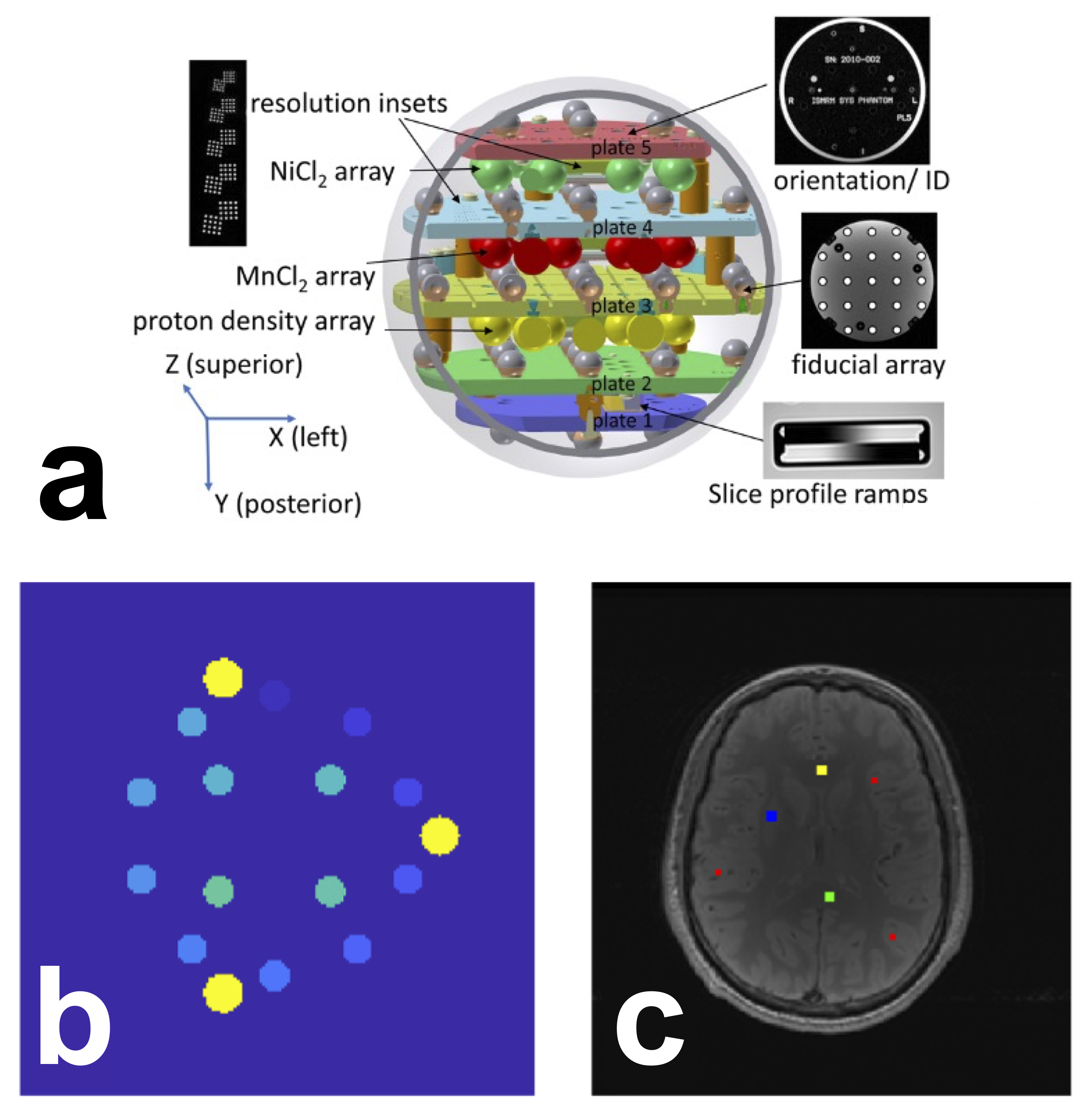
A schematic of the phantom is shown in Figure 1-a. The T1 plate (NiCl2 array) of the phantom has 14 spheres that were labeled as the regions-of-interest (ROI) using a numerical mask template created in MATLAB, provided by NIST researchers (Figure 1-b). To avoid potential edge effects in the T1 maps, the ROI labels were reduced to 60% of the expected sphere diameter. A registration pipeline in Python using the Advanced Normalization Tools (ANTs) [Avants et al., 2009] was developed and shared in the analysis repository of our GitHub organization (https://github.com/rrsg2020/analysis, filename: register_t1maps_nist.py, commit ID: 8d38644). Briefly, a label-based registration was first applied to obtain a coarse alignment, followed by an affine registration (gradientStep: 0.1, metric: cross correlation, number of steps: 3, iterations: 100/100/100, smoothness: 0/0/0, sub-sampling: 4/2/1) and a BSplineSyN registration (gradientStep:0.5, meshSizeAtBaseLevel:3, number of steps: 3, iterations: 50/50/10, smoothness: 0/0/0, sub-sampling: 4/2/1). The ROI labels template was nonlinearly registered to each T1 map uploaded to OSF.
Figure 1 Schematic of the system phantom (a) used in this challenge. Reproduced and cropped from Stupic et al. 2021 (Stupic et al. 2021) (Creative Commons CC BY license). ROI selection for the ISMRM/NIST phantom (b) and the human brain (c). b) The 14 phantom ROIs (shades of blue/green) were automatically generated using a script provided by NIST. In yellow are the three reference pins in the phantom, i.e. these are not ROIs or spheres. c) Human brain ROIs were manually segmented in four regions: the genu (yellow, 5⨯5 voxels), splenium (green, 5⨯5 voxels), deep gray matter (blue, 5⨯5 voxels), and cortical gray matter (red, three sets of 3⨯3 voxels). Note: due to differences in slice positioning from the single-slice datasets provided by certain sites, for some datasets it was not possible to manually segment an ROI in the genu or deep gray matter. In the case of the missing genu, left or right frontal white matter (WM) was selected; for deep gray matter (GM), it was omitted entirely for those cases.
Manual ROIs were segmented by a single researcher (M.B., 11+ years of neuroimaging experience) using FSLeyes [McCarthy, 2022] in four regions for the human datasets Figure 1-c): located in the genu, splenium, deep gray matter, and cortical gray matter. Automatic segmentation was not used because the data were single-slice and there was inconsistent slice positioning between datasets.
2.4.3 | Analysis and Statistics#
Analysis code and scripts were developed and shared in a version-tracked public GitHub repository 7. T1 fitting and main data analysis was performed for all datasets by one of the challenge organizers (M.B.). Python-based Jupyter Notebooks were used for both the quality assurance and main analysis workflows. The computational environment requirements were containerized in Docker [Boettiger, 2015, Merkel, 2014], allowing for an executable environment that can reproduce the analysis in a web browser through MyBinder 8 [Project Jupyter et al., 2018]. Python scripts handled reference data, database handling, ROI masking, and general analysis tools, while configuration files managed the dataset information which were downloaded and pooled using a script (make_pooled_datasets.py). The databases were created using a reproducible Jupyter Notebook and subsequently saved in the repository.
For the ISMRM/NIST phantom data, mean T1 values for each ROI were compared with temperature-corrected reference values and visualized in three different types of plots (linear axes, log-log axes, and error relative to the reference value). This comparison was repeated for individual measurements at each site and for all measurements grouped together. Temperature correction was carried out via nonlinear interpolation 9 of the set of reference NIST T1 values between 16 °C and 26 °C (2 °C intervals), listed in the phantom technical specifications. For the human datasets, a notebook was created to plot the mean and standard deviations for each tissue ROI from all submissions from all sites. All quality assurance and analysis plot images were saved to the repository for ease-of-access and a timestamped version-controlled record of the state of the analysis figures. The database files of ROI values and acquisition details for all submissions were also saved to the repository.
An interactive dashboard 10 was developed in Dash by Plotly (Plotly Technologies Inc. 2015) and hosted by NeuroLibre [Karakuzu et al., 2022] to enable real-time exploration of the data, analysis, and statistics of the challenge results. The dashboard reports descriptive statistics for a variety of alternative looks at phantom and brain data, as well as some statistical comparisons (e.g., the hierarchical shift function 11). The data was collected from the pre-prepared databases of masked ROI values and incorporated other database information, such as phantom version, temperature, MRI system, and reference values. The interactive dashboard displays these results for all measurements at all sites.
3 | RESULTS#
3.1 | Dashboard#
To disseminate the challenge results, a web-based dashboard was developed (Figure 2, https://rrsg2020.dashboards.neurolibre.org). The landing page (Figure 2-a) showcases the relationship between the phantom and brain datasets acquired at different sites/vendors. Navigating to the phantom section leads to the information about the submitted datasets, such as the mean/std/median/CoV for each sphere, % difference from the reference values, number of scans, and temperature (Figure 2-b, left). Other options allow users to limit the results by specific versions of the phantom or the MRI manufacturer. Selecting either “By Sphere” (Figure 2-b, right) or “By Site” tabs will display whisker plots for the selected options, enabling further exploration of the datasets.
Returning to the home page and selecting the brain section allows exploration of information on the brain datasets (Figure 2-c, left), such as mean T1 and STD for different ROI regions, as well as selection of specific MRI manufacturers. Choosing the “By Regions” tab provides whisker plots of the datasets for the selected ROI (Figure 2-c, right), similar to the plots for the phantom.
Figure 2 Dashboard. a) welcome page listing all the sites, the types of subject, and scanner, and the relationship between the three. Row b) shows two of the phantom dashboard tabs, and row c) shows two of the human data dashboard tabs Link: https://rrsg2020.dashboards.neurolibre.org
3.2 | Submissions#
Eighteen submissions were included in the analysis, which resulted in 38 T1 maps of the NIST/system phantom, and 56 brain T1 maps. Figure 3 illustrates all the submissions that acquired phantom data (Figure 3-a) and human data (Figure 3-b), the number of scanners used for each submission, and the number of T1 mapping datasets. It should be noted that these numbers include a subset of measurements where both complex and magnitude-only data from the same acquisition were used to fit T1 maps, thus the total number of unique acquisitions is lower than the numbers reported above (27 for phantom data and 44 for human data). The datasets were collected on three MRI manufacturers (Siemens, GE, Philips) and were acquired at 3T 12, except for one dataset acquired at 0.35T (the ViewRay MRidian MR-linac) . To showcase the heterogeneity of the actual T1 map data from the independently-implemented submissions, Figure 4 displays six T1 maps of the phantoms submitted to the challenge.
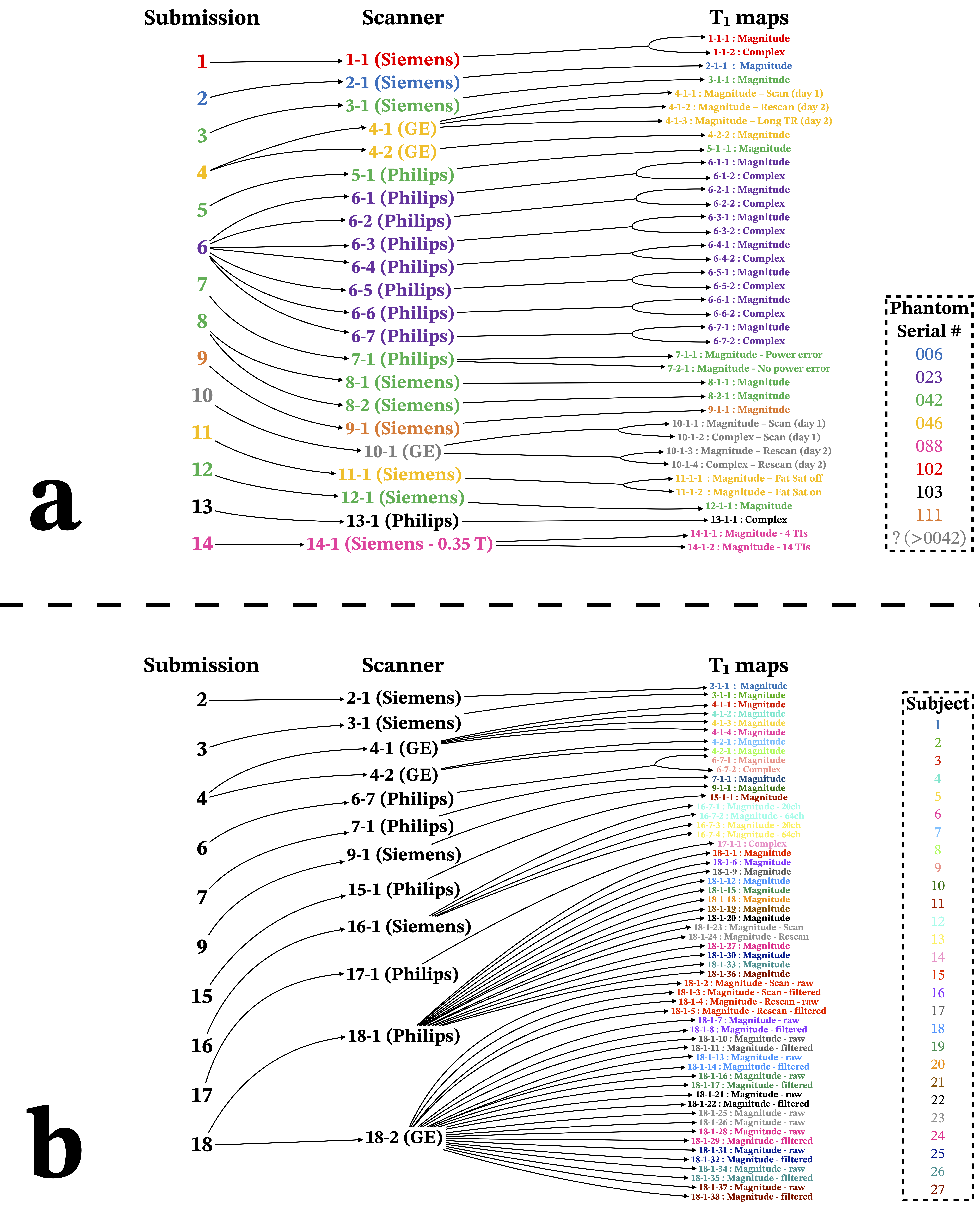
Figure 3 Complete list of the datasets submitted to the challenge. Submissions that included phantom data are shown in a), and those that included human brain data are shown in b). Submissions were assigned numbers to keep track of which submissions included both phantom and human data. Some submissions included datasets acquired on multiple scanners. For the phantom (panel a), each submission acquired all their data using a single phantom, however some researchers shared the same physical phantom with each other (same color). Some additional details about the datasets are included in the T1 maps column, if relevant. Note that for complex datasets in the magnitude/phase format, T1 maps were calculated both using magnitude-only data and complex-data, but these were from the same measurement (branching off arrow).
Of these datasets, several submissions went beyond the minimum acquisition and acquired additional datasets using the ISMRM/NIST phantom, such as a traveling phantom (7 scanners/sites), scan-rescan, same-day rescans on two MRIs, short TR vs long TR, and 4 point TI vs 14 point TI. For humans, one site acquired 13 subjects on two scanners (different manufacturers), one site acquired 6 subjects, and one site acquired a subject using two different head coils (20 channels vs. 64 channels).
from os import path
from pathlib import Path
import os
from repo2data.repo2data import Repo2Data
if build == 'latest':
if path.isdir('analysis')== False:
!git clone https://github.com/rrsg2020/analysis.git
dir_name = 'analysis'
analysis = os.listdir(dir_name)
for item in analysis:
if item.endswith(".ipynb"):
os.remove(os.path.join(dir_name, item))
if item.endswith(".md"):
os.remove(os.path.join(dir_name, item))
elif build == 'archive':
if os.path.isdir(Path('../../data')):
data_path = ['../../data/rrsg-2020-neurolibre']
else:
# define data requirement path
data_req_path = os.path.join("..", "binder", "data_requirement.json")
# download data
repo2data = Repo2Data(data_req_path)
data_path = repo2data.install()
# Imports
from pathlib import Path
import pandas as pd
import json
import nibabel as nib
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.animation as animation
from IPython.display import Video
import glob
from analysis.src.plots import *
from analysis.make_pooled_datasets import *
# Configurations
if build == 'latest':
configFile = Path('analysis/configs/3T_NIST_T1maps.json')
data_folder_name = 'analysis/3T_NIST_T1maps'
configFile_raw = Path('analysis/configs/3T_NIST.json')
data_folder_name_raw = 'analysis/3T_NIST'
elif build=='archive':
configFile = Path(data_path[0] + '/analysis/configs/3T_NIST_T1maps.json')
data_folder_name = data_path[0] + '/analysis/3T_NIST_T1maps'
configFile_raw = Path(data_path[0] + '/analysis/configs/3T_NIST.json')
data_folder_name_raw = data_path[0] + '/analysis/3T_NIST'
# Download datasets
if not Path(data_folder_name).exists():
print(Path(data_folder_name))
make_pooled_dataset(configFile, data_folder_name)
if not Path(data_folder_name_raw).exists():
make_pooled_dataset(configFile_raw, data_folder_name_raw)
with open(configFile) as json_file:
configJson = json.load(json_file)
with open(configFile_raw) as json_file:
configJson_raw = json.load(json_file)
def get_image(dataset_name, key2):
# Load T1 image data
t1_file = configJson[dataset_name]['datasets'][key2]['imagePath']
t1 = nib.load(Path(data_folder_name) / t1_file)
t1_volume = t1.get_fdata()
# Load raw image data
raw_file = configJson_raw[dataset_name]['datasets'][key2]['imagePath']
raw = nib.load(Path(data_folder_name_raw) / raw_file)
raw_volume = raw.get_fdata()
# Handle 2D vs 3D volume case
dims = t1_volume.shape
if (len(dims) == 2) or (np.min(dims) == 1):
im = np.rot90(t1_volume)
TI_1 = np.rot90(np.squeeze(raw_volume[:,:,0,0]))
TI_2 = np.rot90(np.squeeze(raw_volume[:,:,0,1]))
TI_3 = np.rot90(np.squeeze(raw_volume[:,:,0,2]))
TI_4 = np.rot90(np.squeeze(raw_volume[:,:,0,3]))
else:
index_smallest_dim = np.argmin(dims)
numberOfSlices = dims[index_smallest_dim]
midSlice = int(np.round(numberOfSlices/2))
if index_smallest_dim == 0:
im = np.rot90(np.squeeze(t1_volume[midSlice,:,:]))
TI_1 = np.rot90(np.squeeze(raw_volume[midSlice,:,:,0]))
TI_2 = np.rot90(np.squeeze(raw_volume[midSlice,:,:,1]))
TI_3 = np.rot90(np.squeeze(raw_volume[midSlice,:,:,2]))
TI_4 = np.rot90(np.squeeze(raw_volume[midSlice,:,:,3]))
elif index_smallest_dim == 1:
im = np.rot90(np.squeeze(t1_volume[:,midSlice,:]))
TI_1 = np.rot90(np.squeeze(raw_volume[:,midSlice,:,0]))
TI_2 = np.rot90(np.squeeze(raw_volume[:,midSlice,:,1]))
TI_3 = np.rot90(np.squeeze(raw_volume[:,midSlice,:,2]))
TI_4 = np.rot90(np.squeeze(raw_volume[:,midSlice,:,3]))
elif index_smallest_dim == 2:
im = np.rot90(np.squeeze(t1_volume[:,:,midSlice]))
TI_1 = np.rot90(np.squeeze(raw_volume[:,:,midSlice,0]))
TI_2 = np.rot90(np.squeeze(raw_volume[:,:,midSlice,1]))
TI_3 = np.rot90(np.squeeze(raw_volume[:,:,midSlice,2]))
TI_4 = np.rot90(np.squeeze(raw_volume[:,:,midSlice,3]))
xAxis = np.linspace(0,im.shape[0]-1, num=im.shape[0])
yAxis = np.linspace(0,im.shape[1]-1, num=im.shape[1])
return im, xAxis, yAxis, TI_1, TI_2, TI_3, TI_4
im_1, xAxis_1, yAxis_1, TI_1_1, TI_2_1, TI_3_1, TI_4_1 = get_image('wang_MDAnderson_NIST', 'day2_mag')
im_2, xAxis_2, yAxis_2, TI_1_2, TI_2_2, TI_3_2, TI_4_2 = get_image('CStehningPhilipsClinicalScienceGermany_NIST', 'Bonn_MR1_magnitude')
im_3, xAxis_3, yAxis_3, TI_1_3, TI_2_3, TI_3_3, TI_4_3 = get_image('mrel_usc_NIST', 'Session1_MR1')
im_4, xAxis_4, yAxis_4, TI_1_4, TI_2_4, TI_3_4, TI_4_4 = get_image('karakuzu_polymtl_NIST', 'mni')
im_5, xAxis_5, yAxis_5, TI_1_5, TI_2_5, TI_3_5, TI_4_5 = get_image('madelinecarr_lha_NIST', 'one')
im_6, xAxis_6, yAxis_6, TI_1_6, TI_2_6, TI_3_6, TI_4_6 = get_image('matthewgrechsollars_ICL_NIST', 'magnitude')
im_6 = np.flipud(im_6)
TI_1_6 = np.flipud(TI_1_6)
TI_2_6 = np.flipud(TI_2_6)
TI_3_6 = np.flipud(TI_3_6)
TI_4_6 = np.flipud(TI_4_6)
# PYTHON CODE
# Module imports
import matplotlib.pyplot as plt
from PIL import Image
from matplotlib.image import imread
import scipy.io
import plotly.graph_objs as go
import numpy as np
from plotly import __version__
from plotly.offline import init_notebook_mode, iplot, plot
config={'showLink': False, 'displayModeBar': False, 'responsive': True}
init_notebook_mode(connected=True)
from IPython.display import display, HTML
import os
import markdown
import random
from scipy.integrate import quad
import warnings
warnings.filterwarnings('ignore')
xAxis = np.linspace(0,256*3-1, num=256*3)
yAxis = np.linspace(0,256*2-1, num=256*2)
# T1 maps
im_2_padded = np.pad(im_2,32)
images_1 = np.concatenate((im_1, im_5, im_3), axis=1)
images_2 = np.concatenate((im_4, im_2_padded, im_6), axis=1)
images = np.concatenate((images_2, images_1), axis=0)
# TI_1 maps
TI_1_2_padded = np.pad(TI_1_2,32)
TI_1_images_1 = np.concatenate((TI_1_1, TI_1_5, TI_1_3), axis=1)
TI_1_images_2 = np.concatenate((TI_1_4, TI_1_2_padded, TI_1_6), axis=1)
TI_1_images = np.concatenate((TI_1_images_2, TI_1_images_1), axis=0)
# TI_2 maps
TI_2_2_padded = np.pad(TI_2_2,32)
TI_2_images_1 = np.concatenate((TI_2_1, TI_2_5, TI_2_3), axis=1)
TI_2_images_2 = np.concatenate((TI_2_4, TI_2_2_padded, TI_2_6), axis=1)
TI_2_images = np.concatenate((TI_2_images_2, TI_2_images_1), axis=0)
# TI_3 maps
TI_3_2_padded = np.pad(TI_3_2,32)
TI_3_images_1 = np.concatenate((TI_3_1, TI_3_5, TI_3_3), axis=1)
TI_3_images_2 = np.concatenate((TI_3_4, TI_3_2_padded, TI_3_6), axis=1)
TI_3_images = np.concatenate((TI_3_images_2, TI_3_images_1), axis=0)
# TI_4 maps
TI_4_2_padded = np.pad(TI_4_2,32)
TI_4_images_1 = np.concatenate((TI_4_1, TI_4_5, TI_4_3), axis=1)
TI_4_images_2 = np.concatenate((TI_4_4, TI_4_2_padded, TI_4_6), axis=1)
TI_4_images = np.concatenate((TI_4_images_2, TI_4_images_1), axis=0)
trace1 = go.Heatmap(x = xAxis,
y = yAxis_1,
z=images,
zmin=0,
zmax=3000,
colorscale='viridis',
colorbar={"title": 'T<sub>1</sub> (ms)',
'titlefont': dict(
family='Times New Roman',
size=26,
)
},
xaxis='x2',
yaxis='y2',
visible=True)
trace2 = go.Heatmap(x = xAxis,
y = yAxis_1,
z=TI_1_images,
zmin=0,
zmax=3000,
colorscale='gray',
colorbar={"title": 'T<sub>1</sub> (ms)',
'titlefont': dict(
family='Times New Roman',
size=26,
color='white'
)
},
xaxis='x2',
yaxis='y2',
visible=False)
trace3 = go.Heatmap(x = xAxis,
y = yAxis_1,
z=TI_2_images,
zmin=0,
zmax=3000,
colorscale='gray',
colorbar={"title": 'T<sub>1</sub> (ms)',
'titlefont': dict(
family='Times New Roman',
size=26,
color='white'
)
},
xaxis='x2',
yaxis='y2',
visible=False)
trace4 = go.Heatmap(x = xAxis,
y = yAxis_1,
z=TI_3_images,
zmin=0,
zmax=3000,
colorscale='gray',
colorbar={"title": 'T<sub>1</sub> (ms)',
'titlefont': dict(
family='Times New Roman',
size=26,
color='white'
)
},
xaxis='x2',
yaxis='y2',
visible=False)
trace5 = go.Heatmap(x = xAxis,
y = yAxis_1,
z=TI_4_images,
zmin=0,
zmax=3000,
colorscale='gray',
colorbar={"title": 'T<sub>1</sub> (ms)',
'titlefont': dict(
family='Times New Roman',
size=26,
color='white'
)
},
xaxis='x2',
yaxis='y2',
visible=False)
data=[trace1, trace2, trace3, trace4, trace5]
updatemenus = list([
dict(active=0,
x = 0.4,
xanchor = 'left',
y = -0.15,
yanchor = 'bottom',
direction = 'up',
font=dict(
family='Times New Roman',
size=16
),
buttons=list([
dict(label = 'T<sub>1</sub> maps',
method = 'update',
args = [{'visible': [True, False, False, False, False],
'showscale': True,},
]),
dict(label = 'TI = 50 ms',
method = 'update',
args = [
{
'visible': [False, True, False, False, False],
'showscale': True,},
]),
dict(label = 'TI = 400 ms',
method = 'update',
args = [{'visible': [False, False, True, False, False],
'showscale': True,},
]),
dict(label = 'TI = 1100 ms',
method = 'update',
args = [{'visible': [False, False, False, True, False],
'showscale': True,},
]),
dict(label = 'TI ~ 2500 ms',
method = 'update',
args = [{'visible': [False, False, False, False, True],
'showscale': True,},
]),
])
)
])
layout = dict(
width=960,
height=649,
margin = dict(
t=40,
r=50,
b=10,
l=50),
xaxis = dict(range = [0,256*3-1], autorange = False,
showgrid = False, zeroline = False, showticklabels = False,
ticks = '', domain=[0, 1]),
yaxis = dict(range = [0,256*2-1], autorange = False,
showgrid = False, zeroline = False, showticklabels = False,
ticks = '', domain=[0, 1]),
xaxis2 = dict(range = [0,256*3-1], autorange = False,
showgrid = False, zeroline = False, showticklabels = False,
ticks = '', domain=[0, 1]),
yaxis2 = dict(range = [0,256*2-1], autorange = False,
showgrid = False, zeroline = False, showticklabels = False,
ticks = '', domain=[0, 1], anchor='x2'),
showlegend = False,
autosize = False,
updatemenus=updatemenus
)
fig = dict(data=data, layout=layout)
#iplot(fig, filename = 'basic-heatmap', config = config)
plot(fig, filename = 'figure2.html', config = config)
display(HTML('figure2.html'))