How to add new bundles into pyAFQ (Optic Radiations Example)
pyAFQ is designed to be customizable and extensible. This example shows how you can customize it to define a new bundle based on a definition of waypoint and endpoint ROIs of your design.
In this case, we add the optic radiations, based on work by Caffara et al. The optic radiations (OR) are the primary projection of visual information from the lateral geniculate nucleus of the thalamus to the primary visual cortex. Studying the optic radiations with dMRI provides a linkage between white matter tissue properties, visual perception and behavior, and physiological responses of the visual cortex to visual stimulation.
from paths import afq_home
import os.path as op
import plotly
import numpy as np
from IPython.display import Image
from AFQ.api.group import GroupAFQ
import AFQ.api.bundle_dict as abd
import AFQ.data.fetch as afd
from AFQ.definitions.image import ImageFile, RoiImage
np.random.seed(1234)1Get dMRI data¶
We will analyze one subject from the Healthy Brain Network Processed Open Diffusion Derivatives dataset (HBN-POD2). We’ll use a fetcher to get preprocessed dMRI data for one of the >2,000 subjects in that study.
study_dir = afd.fetch_hbn_preproc(["NDARAA948VFH"])[1]2Define custom BundleDict object¶
The BundleDict object holds information about “include” and “exclude” ROIs,
as well as endpoint ROIs, and whether the bundle crosses the midline.
or_rois = afd.read_or_templates()
bundles = abd.OR_bd()3Define GroupAFQ object¶
For tractography, we use CSD-based probabilistic tractography seeding
extensively (n_seeds=4 means 81 seeds per voxel!), but only within the ROIs.
brain_mask_definition = ImageFile(
suffix="mask",
filters={'desc': 'brain',
'space': 'T1w',
'scope': 'qsiprep'})
my_afq = GroupAFQ(
bids_path=study_dir,
preproc_pipeline="qsiprep",
participant_labels=["NDARAA948VFH"],
output_dir=op.join(study_dir, "derivatives", "afq_or"),
brain_mask_definition=brain_mask_definition,
tracking_params={"n_seeds": 4,
"directions": "prob",
"odf_model": "CSD",
"seed_mask": RoiImage()},
bundle_info=bundles)
my_afq.export("profiles")INFO:AFQ:No stop mask given, using FA (or first scalar if none are FA)thresholded to 0.2
{'NDARAA948VFH': '/home/jovyan/data/tractometry/tractometry/HBN/derivatives/afq_or/sub-NDARAA948VFH/ses-HBNsiteRU/dwi/sub-NDARAA948VFH_ses-HBNsiteRU_acq-64dir_desc-profiles_tractography.csv'}4Visualize a montage¶
One way to examine the output of the pyAFQ pipeline is by creating a montage of images of a particular bundle across a group of participants.
Image(filename=op.join(
afq_home,
"HBN/derivatives/afq_or/",
"bundle-Left Optic Radiation_view-Axial_idx-0_montage.png"))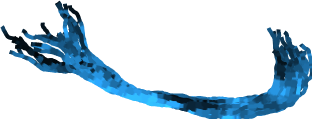
5Interactive bundle visualization¶
Another way to examine the outputs is to export the bundles as interactive HTML files.
bundle_html = my_afq.export("all_bundles_figure")
plotly.io.show(bundle_html["NDARAA948VFH"][0])6References¶
Caffarra S, Joo SJ, Bloom D, Kruper J, Rokem A, Yeatman JD. Development of the visual white matter pathways mediates development of electrophysiological responses in visual cortex. Hum Brain Mapp. 2021. Caffarra et al. (2021)
Caffarra S, Kanopka K, Kruper J, Richie-Halford A, Roy E, Rokem A, Yeatman JD. Development of the alpha rhythm is linked to visual white matter pathways and visual detection performance. bioRxiv. Caffarra et al. (2023)
Alexander LM, Escalera J, Ai L, et al. An open resource for transdiagnostic research in pediatric mental health and learning disorders. Sci Data. 2017. Alexander et al. (2017)
Richie-Halford A, Cieslak M, Ai L, et al. An analysis-ready and quality controlled resource for pediatric brain white-matter research. Scientific Data. 2022. Richie-Halford et al. (2022)
Cieslak M, Cook PA, He X, et al. QSIPrep: an integrative platform for preprocessing and reconstructing diffusion MRI data. Nat Methods. 2021. Cieslak et al. (2021)
- Caffarra, S., Joo, S. J., Bloom, D., Kruper, J., Rokem, A., & Yeatman, J. D. (2021). Development of the visual white matter pathways mediates development of electrophysiological responses in visual cortex. Human Brain Mapping, 42(17), 5785–5797. 10.1002/hbm.25654
- Caffarra, S., Kanopka, K., Kruper, J., Richie-Halford, A., Roy, E., Rokem, A., & Yeatman, J. D. (2023). Development of the Alpha Rhythm Is Linked to Visual White Matter Pathways and Visual Detection Performance. The Journal of Neuroscience, 44(6), e0684232023. 10.1523/jneurosci.0684-23.2023
- Alexander, L. M., Escalera, J., Ai, L., Andreotti, C., Febre, K., Mangone, A., Vega-Potler, N., Langer, N., Alexander, A., Kovacs, M., Litke, S., O’Hagan, B., Andersen, J., Bronstein, B., Bui, A., Bushey, M., Butler, H., Castagna, V., Camacho, N., … Milham, M. P. (2017). An open resource for transdiagnostic research in pediatric mental health and learning disorders. Scientific Data, 4(1). 10.1038/sdata.2017.181
- Richie-Halford, A., Cieslak, M., Ai, L., Caffarra, S., Covitz, S., Franco, A. R., Karipidis, I. I., Kruper, J., Milham, M., Avelar-Pereira, B., Roy, E., Sydnor, V. J., Yeatman, J. D., Abbott, N. J., Anderson, J. A. E., Gagana, B., Bleile, M., Bloomfield, P. S., Bottom, V., … Rokem, A. (2022). An analysis-ready and quality controlled resource for pediatric brain white-matter research. Scientific Data, 9(1). 10.1038/s41597-022-01695-7
- Cieslak, M., Cook, P. A., He, X., Yeh, F.-C., Dhollander, T., Adebimpe, A., Aguirre, G. K., Bassett, D. S., Betzel, R. F., Bourque, J., Cabral, L. M., Davatzikos, C., Detre, J. A., Earl, E., Elliott, M. A., Fadnavis, S., Fair, D. A., Foran, W., Fotiadis, P., … Satterthwaite, T. D. (2021). QSIPrep: an integrative platform for preprocessing and reconstructing diffusion MRI data. Nature Methods, 18(7), 775–778. 10.1038/s41592-021-01185-5