How to add new bundles into pyAFQ (SLF 1/2/3 Example)
pyAFQ is designed to be customizable and extensible. This example shows how you can customize it to define a new bundle based on a definition of waypoint and endpoint ROIs of your design.
In this case, we add sub-bundles of the superior longitudinal fasciculus, based on work by Sami et al. 2024
from paths import afq_homeimport os.path as op
import plotly
import numpy as np
from IPython.display import Image
from AFQ.api.group import GroupAFQ
import AFQ.api.bundle_dict as abd
import AFQ.data.fetch as afd
from AFQ.definitions.image import ImageFile, RoiImage
np.random.seed(1234)1Get dMRI data¶
We will analyze twelve subjects from the Healthy Brain Network Processed Open Diffusion Derivatives dataset (HBN-POD2) Richie-Halford et al. (2022).
The data gets organized into a BIDS-compatible format.
_, study_dir = afd.fetch_hbn_preproc(['NDARAA948VFH'])2Define custom BundleDict object¶
We define the bundles with include/exclude ROIs and specific cleaning parameters.
template_dir = op.join(afq_home, 'SLF_ROIs')
templates_afq = afd.read_templates(as_img=False)
# You can provide paths to the ROIs in the "include" and "exclude" keys of the bundle definition
bundles = abd.BundleDict({
"L_SLF1": {
"include": [template_dir + '/SFgL.nii.gz', template_dir + '/PaL.nii.gz'],
"exclude": [templates_afq['SLFt_roi2_L']],
"cross_midline": False,
"mahal": {"clean_rounds": 20, "length_threshold": 4, "distance_threshold": 2}
}
})3Define GroupAFQ object¶
We configure the AFQ pipeline with predefined parameters and start the process.
brain_mask_definition = ImageFile(
suffix="mask",
filters={'desc': 'brain', 'space': 'T1w', 'scope': 'qsiprep'}
)
my_afq = GroupAFQ(
bids_path=study_dir,
preproc_pipeline="qsiprep",
participant_labels=["NDARAA948VFH"],
output_dir=op.join(study_dir, "derivatives", "afq_slf"),
brain_mask_definition=brain_mask_definition,
tracking_params={"n_seeds": 4, "directions": "prob", "odf_model": "CSD", "seed_mask": RoiImage()},
segmentation_params={"parallel_segmentation": {"engine": "serial"}},
bundle_info=bundles
)
my_afq.export("profiles")INFO:AFQ:No stop mask given, using FA (or first scalar if none are FA)thresholded to 0.2
{'NDARAA948VFH': '/home/jovyan/data/tractometry/tractometry/HBN/derivatives/afq_slf/sub-NDARAA948VFH/ses-HBNsiteRU/dwi/sub-NDARAA948VFH_ses-HBNsiteRU_acq-64dir_desc-profiles_tractography.csv'}4Visualize a montage¶
We create montage visualizations of the SLF bundles.
montage = my_afq.group_montage("L_SLF1", (3, 4), "Sagittal", "left", slice_pos=0.5)
Image(filename=montage[0])INFO:AFQ:Generating Montage...
0%| | 0/1 [00:00<?, ?it/s]INFO:AFQ:Loading Volume...
INFO:AFQ:Loading Volume...
INFO:AFQ:Loading Stateful Tractogram...
/srv/conda/envs/notebook/lib/python3.11/site-packages/AFQ/utils/streamlines.py:93: UserWarning:
Pass ['to_space'] as keyword args. From version 2.0.0 passing these as positional arguments will result in an error.
INFO:AFQ:Generating colorful lines from tractography...
100%|██████████| 1/1 [00:06<00:00, 6.77s/it]100%|██████████| 1/1 [00:06<00:00, 6.78s/it]
INFO:AFQ:Montage file already exists, skipping.
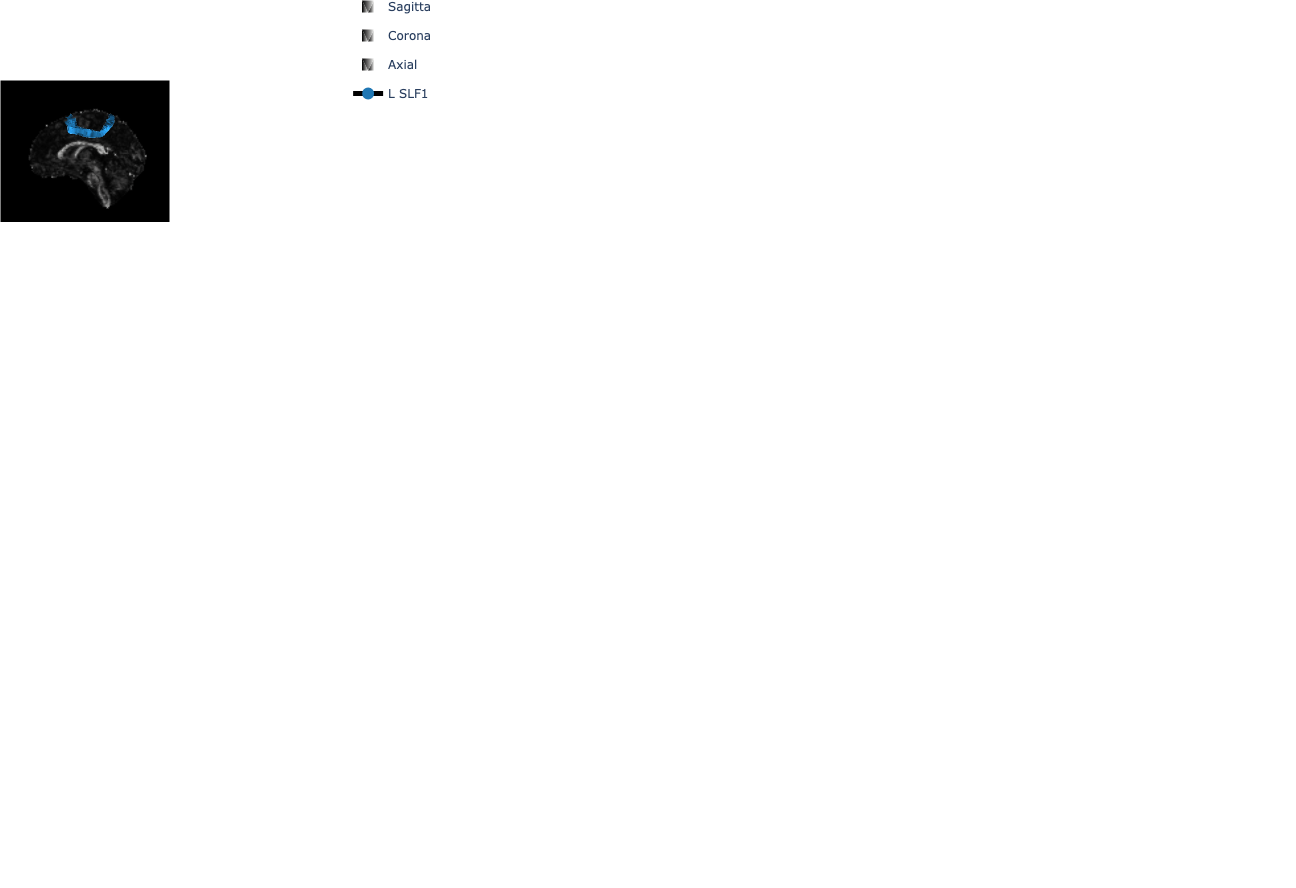
5Interactive bundle visualization¶
We export individual bundle figures as interactive HTML files.
bundle_html = my_afq.export("all_bundles_figure")
plotly.io.show(bundle_html["NDARAA948VFH"][0])6References¶
Richie-Halford A, Cieslak M, Ai L, et al. An analysis-ready and quality controlled resource for pediatric brain white-matter research. Scientific Data. 2022. Richie-Halford et al. (2022)
Cieslak M, Cook PA, He X, et al. QSIPrep: an integrative platform for preprocessing and reconstructing diffusion MRI data. Nat Methods. 2021. Cieslak et al. (2021).
- Sagi, R., Taylor, J. S. H., Neophytou, K., Cohen, T., Rapp, B., Rastle, K., & Ben-Shachar, M. (2024). White matter associations with spelling performance. Brain Structure and Function, 229(9), 2115–2135. 10.1007/s00429-024-02775-7
- Richie-Halford, A., Cieslak, M., Ai, L., Caffarra, S., Covitz, S., Franco, A. R., Karipidis, I. I., Kruper, J., Milham, M., Avelar-Pereira, B., Roy, E., Sydnor, V. J., Yeatman, J. D., Abbott, N. J., Anderson, J. A. E., Gagana, B., Bleile, M., Bloomfield, P. S., Bottom, V., … Rokem, A. (2022). An analysis-ready and quality controlled resource for pediatric brain white-matter research. Scientific Data, 9(1). 10.1038/s41597-022-01695-7
- Cieslak, M., Cook, P. A., He, X., Yeh, F.-C., Dhollander, T., Adebimpe, A., Aguirre, G. K., Bassett, D. S., Betzel, R. F., Bourque, J., Cabral, L. M., Davatzikos, C., Detre, J. A., Earl, E., Elliott, M. A., Fadnavis, S., Fair, D. A., Foran, W., Fotiadis, P., … Satterthwaite, T. D. (2021). QSIPrep: an integrative platform for preprocessing and reconstructing diffusion MRI data. Nature Methods, 18(7), 775–778. 10.1038/s41592-021-01185-5